We have recently been using Bowtie2 for sequence comparisons related to shotgun metagenomics, where we directly sequence samples that contain mixtures of DNA from different organisms, with no PCR. Bowtie2 alignments can be made very stringently (>90% identity), and computed very rapidly for large data files with hundreds of millions of sequence reads. This allows us to identify DNA fragments by species and by gene function, provided we have a well-annotated database that contains DNA sequences from similar microbes. I know that MetaPhlan and other tools already do this, but I want to focus on the difference between bacteria (and viruses) at the strain level.
I have been playing around with the idea of constructing a database that will give strain-specific identification for human-associated microbes using Bowtie2 alignments with shotgun metagenomic data. There are plenty of sources for bacterial genome sequences, but the PATRIC database has a very good collection of human pathogen genomes. However the database is very redundant - with dozens or even hundreds of different strains for the same species, and the whole thing is too large to build into a Bowtie database (>350 Gb). So I am looking at ways to eliminate redundancy from the database while retaining high sensitivity at the species level to identify any sequence fragment, not just specific marker genes, and the ability to make strain-specific alignments. So I want to identify which genome fragments have shared sequences among a group of strains within a single species and which fragments are unique to each strain.
Since our sequence reads are usually ~100 bases, We are looking the similarity between bacterial strains when the genome is chopped into 100 bp pieces.
Bowtie2 can be limited to only perfect alignments (100 bases with zero mismatches) using the parameters --end-to-end and --scoremin C, 0, -1 and to something similar to 99% identity with --scoremin ‘L,0, -0.06’
Between two strains of Streptococcus agalactiae, if we limit the Bowtie2 alignments to perfect matches, half of the fragments align. So at the 100 base level, half of the genome is identical, and the other half has at least one variant. At 99% similarity (one mismatch per read), about 2/3 of the fragments align, and the other 1/3 has more than one variant, or in some cases no-similarity at all to the other genome. Yuhan Hao extended this experiment to another species (Strep pyogenes), where we see almost no alignment to Strep ag. of genome fragments at 100%, 99%, or ~97% identity (see the Table below). I have previously used the 97% sequence identity threshold to separate bacterial species, but I thought it was only for 16S rDNA sequences - here it seems to apply to almost every 100 base fragment in the whole genome.
So we can build a database with one reference genome per species and safely eliminate the 2/3 of fragments that are nearly identical between strains, and retain only those portions of the genome that are unique to each strain. I'm going to think about how to apply this iteratively (without creating a huge computing task), so we add only the truly unique fragments for EACH strain, rather than just testing if a fragment differs from the Reference for that species. With such a database, stringent Bowtie2 alignments will identify each sequence read by species and by strain and have very low false matches across different species.
We can visualize the 100 base alignments between two strains at the 100% identity level, 99% identity, and at the least stringent Bowtie2 setting (very fast local) to see where the strains differ. This makes a pretty picture (below), which reveals some fairly obvious biology: There are parts of the genome that are more conserved and parts that are more variable between strains. The top panel shows only the 100% perfect matches (grey bars), the second panel shows that we can add in some 99% matching reads (white bars with a single vertical color stripe to mark the mismatch base), and the lowest panel shows reads that are highly diverged (lots of colored mismatch bases and some reads that are clearly mis-aligned from elsewhere on the genome). So if we choose only the reads that differ by more than 99%, we can build a database that can use Bowtie2 to identify different strains and have few multiple matches or incorrectly matched reads.
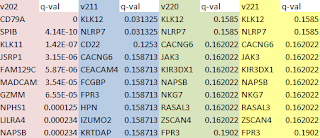
This chart lists the number of UNMATCHED fragments after alignment of all non-overlapping 100 bp fragments from the genomes of each strain to the Strep. agalactiae genome that we arbitrarily chose as the Reference. C 0 -1 corresponds to perfect matches only, L 0 -0.06 is approximately 99% identity (by my calculation), and L 0, -0.2 is about 97% identity. The lower the stringency of alignment, the fewer fragments end up in the "unmatched" file.
S.ag Ref S.ag st2 S.ag
st3 S.pyo st1 S.pyo st2
# 100b kmers 20615 22213 21660 17869 17094
C 0 -1 0 0 11243 10571 17718 16934 (perfect matches)
L 0 -0.06 0 0 6605 5918 17533 16762 (99% ident)
L 0 -0.1 0 0 6534 5839 17533 16762
L 0 -0.12 0 0 4812 4058 17371 16600
L 0 -0.15 0 0 4767 4006 17369 16600
L 0 -0.17 0 0 4756 3997 17369 16600
L 0 -0.2 0 0 4156 3419 17237 16441
(97% ident)