I was working through my RNA-seq tutorial with a group of students this week and they pointed out that my tutorial worksheet was wrong. Cufflinks did not produce any significant DE genes with our test data comparing two small RNA-seq data files. This was surprising to me, since I did the exact same workflow with the same data files last year and it worked out fine. So golly and darn it, I got hit with the irreproducible results bug. We keep past versions of software available on our computing server with an Environment Modules system, so I was able to quickly run a test of the exact same data files (aligned with the same Tophat version to the same reference genome) using different versions of Cufflinks. We have the following versions installed:
cufflinks/2.0.2 (July 2012)
cufflinks/2.1.1 (April 2013)
cufflinks/2.2.0 (March 2014)
cufflinks/2.2.1 (May 2014)
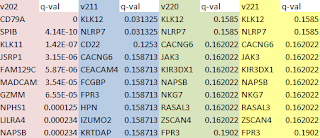
I just used the simple Cuffidiff workflow and looked at the gene_exp.diff output file for each software version. The results are quite different. Version 2.0.2 has 46 genes called "significant=yes" (multi-test adjusted q-value less than 0.05) with q-values running as low as 4.14E-10 (ok, one has a q-value of zero). Now this is not a great result from a biostatistics standpoint, since how can you expect to get significant p-values from RNA expression levels with two samples an no replicates? But it did make for an expedient exercise, since we could take the DE genes into DAVID and look for enriched biological functions and pathways.
Then in version 2.1.1 we have two "significant=yes" genes. In version 2.2.0 we have zero significant genes, and in version 2.2.1 also zero.
The top 10 genes, ranked by q-value also differ. There are no genes in common between the top 10 list for version 2.0.2 and 2.1.1, and only the top 2 genes are shared by version 2.2.0. Thankfully, there are no differences in top genes or q-values between 2.2.0 and 2.2.1 (versions released only 2 months apart). I'm sure that Cole Trapnell et al. are diligently improving the software, but the consequences for those of us trying to use the tools to make some sense out of biology experiments can be unsettling.
5 comments:
Although I agree with you that tool version changes which affect results are frustrating, I'd be more worried about the fact you're trying to infer biology with single replicate data.
In order to get a biologically meaningful result in any study biological replicates are required. All biological system are stochastic to some degree so to be able to discern the real signal over and above the noise, it needs to be measured. That is only possible with replicates. Anything else is flawed.
Even with replicates there can be high variability in gene expression. In the study design process you need to balance statistical power with effect size (e.g. fold change) to determine how many reps are required, although we suggest a minimum of 6 biologial replicates for most experiments. We have a paper in press with RNA on this topic. The preprint is available here: http://arxiv.org/abs/1505.02017
You have completely missed my point. Using the exact same data files and only changing the Cufflinks version, I obtain entirely different ranked lists of differentially regulated genes.
And yes, I do tell my students that you must use replicates for valid studies of gene expression. However, with 15 students in my class (or 45 students in a larger workshop last week), running a full scale Tophat/Cufflinks workflow with ~3 replicates in each of treatment and control conditions would put some serious strain on our server. So instead for the hands-on demo I use just two data files that are artificially small.
You've missed my point. Trying to discern anything meaningful from single-replicate data is futile. I realise the expectation of running different versions of the same tool to generate similar results. There are so many (probably wrong) assumptions with singlicate data that the results are mostly influenced by them rather than the actual data. Just because you can generate results with singlicate data, doesn't mean you should. Also, despite the apparently small increment in version, 2.1.1 is a very different method to 2.0.2 (see the release notes http://cole-trapnell-lab.github.io/cufflinks/releases/v2.1.0/) - it will give very different results.
As an aside, TopHat is known to be notoriously slow, we stopped using it a couple of years ago. Try STAR which is a much faster aligner or try the new alignment-free methods e.g. Sailfish or Kallisto which are very frugal methods. All would be an improvement over TopHat. Note that, although STAR can be used with cufflinks, Kallisto/Sailfish cannot - they require different downstream tools.
My name is Antonio Jefferson Cole, I'm in Salt Lake City, Utah. It is a pleasure for me to write this testimony about how I got healed from genital herpes 12 hours ago. I have read so many comments from some people who have been cured of HIV by Dr. Ekpiku, but I never believed them. I was hurt and depressed, so I was very curious and wanted to try Dr. Ekpiku, so I contacted him on his email Ekpikuspellhomeofgrace@gmail.com when I get in touch with him, he assured me 100% that it will heal me, I begged Him to help me. My treatment was a great success, he healed me as he promised. He sent me his medication and asked me to go for check-up after 2 weeks of taking the medication. I agree with him, I took this medication and went to the check-up after 2 month, to my greatest surprise my result was negative after treatment, I am really happy that I am healed and healthy again. I waited for four months to make sure I was completely healed before writing this testimony. I did another blood test today and was still negative Herpes, so I think its time I tell someone out there with Herpes HSV-1 or HSV-2, HIV, HPV, hepatitis B, diabetes, cancer and so on, Dr. Ekpiku in the email: Ekpikuspellhomeofgrace@hotmail.com . Your phone number is +2348073673757, He is also on whats app. I completely understand how you can feel skeptical about it, because it happened to me, it can be expensive for you, but I'm telling you now that there is a cure for these diseases and I am a living witness firsthand.or you can email me on antoniojeffersoncole@gmail.com for any assistance.
You have completely missed my point. Using the exact same data files and only changing the Cufflinks version, I obtain entirely different ranked lists of differentially regulated genes.
For more information:-
RNA Fixation
RNA Stabilizer
RNA Protect
RNA Sequencing
formaldehyde alternative
Post a Comment